Building an R package is an easy and very convenient way keep your work well organized. Moreover, it facilitates sharing your code with the R community. Here we will discuss about publishing R packages on CRAN and GitHub. The post summarizes the steps required for:
- building and checking a package
- submitting an R package to CRAN
- publishing an R package on GitHub
This post is not meant to be a comprehensive tutorial (you can find detailed information about R packages elsewhere). This post is aimed at summarizing the basic steps that can get you started with R packages. Moreover, I’ll try to highlight how to avoid some of the most common mistakes. I assume you are working on a Linux environment (code was tested on a Ubuntu 14.04 machine.
Building a package using package.skeleton() {utils}
- Clear the workspace and start a new R session
- Load all data you want to be stored in the package (functions and data) in the R session. If you are using R studio, you should see your data and functions in the Global Environment (and nothing else)
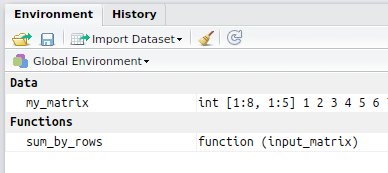
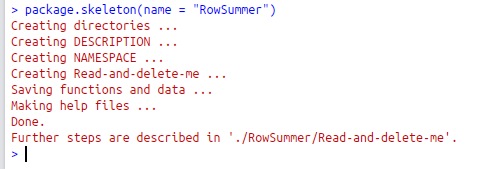
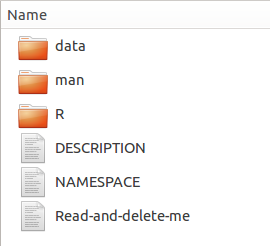
- Call package.skeleton(name = “NameOfYourPackage”). This will create a new folder in the working directory (named according to the name you specified) containing a set of files.
- If you are including one or more dataset that require compression, save it as a compressed file after the package.skeleton() call. You can do so by running save(test, file=”mypackage/data/test.rda”, compress=’xz’)
- Read-and-delete-me file is a reminder of the steps you have to take next. Read it and then delete it.
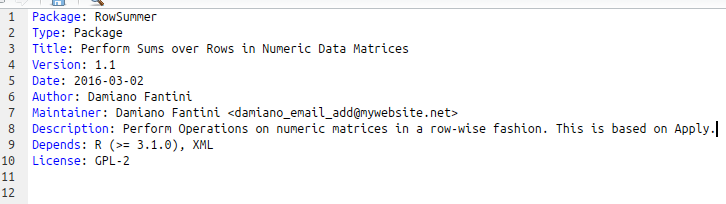
- Open the DESCRIPTION file in RStudio and edit it. In order to comply to CRAN policies, you should:
- write the Title in Title-case (ex.: Perform Operations and Compute Sums on Data Stored in Matrices)
- Description and Title should not start with “This package”, the name of the package or contain useless information (avoid writing that the package works in an R environment or stuff like that)
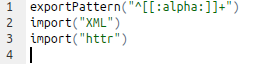
- Imports and Dependencies. Which are the packages your package relies on? Add a “Imports:” or a “Depends:” line in DESCRIPTION
- Specify the type of License. For a list of common CRAN licenses, you can check this link: https://cran.r-project.org/web/licenses/
- An example is shown here.
- Edit the NAMESPACE file in RStudio. It may be a good idea to use roxygen2 for writing documentation and the NAMESPACE. However, if your package is simple, you can easily edit the NAMESPACE manually. A good description of how NAMESPACE works is available here: http://r-pkgs.had.co.nz/namespace.html. Briefly, here you want to tell R where to go and search for external functions used in your package. An example is shown here.
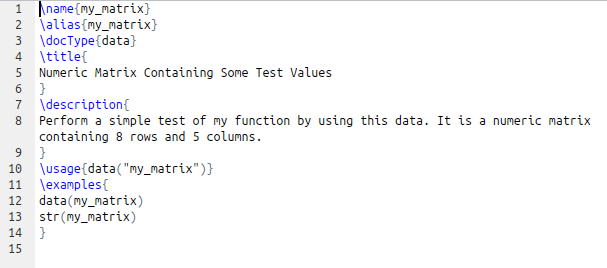
- Open the man folder and edit each .Rd file. These are documentation file that describe each of the objects your package carries (data as well functions). The portions of each document you are supposed to edit are flagged with a comment starting with %% ~~ do something here! ~~. If you don’t need a specific field (ex.: \references{…} ), you can remove it by deleting the whole line.
- Follow the same basic rules described for the DESCRIPTION file
- Edit the documentation file for the package as well
- Carefully describe each argument of each function. It is generally important to state clearly what is the class of each object passed as argument.
- Likewise, it’s highly recommended to describe the type of value(s) returned by each function
- Include examples. Make sure that the data required for running the function is available in the namespace (remember that you can include data objects in the package). Examples runtime should be short. CRAN accepts examples that can complete within 5 seconds. If you want to provide some examples that take longer to execute, place them within a \dontrun { … } declaration. Don’t provide examples that won’t work at all.
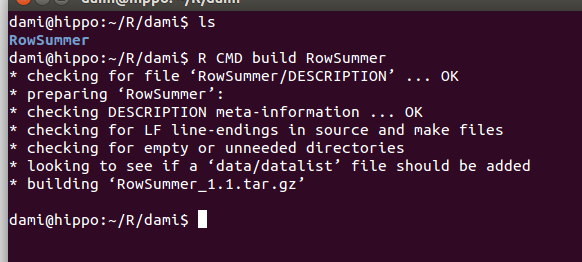
- Once the Documentation files have been edited (remember to remove the Read-and-delete-me file), you can finally build the package. Open a terminal shell, change directory to the current R working directory and then run the following command: R CMD build PackageName
- Congratulations, you just built an R package. If the imports and dependencies were stated correctly and the Documentation was edited without mistakes, the package check should return no errors. Checking a package is important to make sure that the package doesn’t contain mistakes (ex.: mistake in defining the namespace) and that the documentation is correctly written. Before checking a package, you should make sure that:
- latex is installed in your system (http://milq.github.io/install-latex-ubuntu-debian/)
- MASS, spatial and lattice packages are installed in your R system (if a package is missing, you’ll be notified during the check phase).
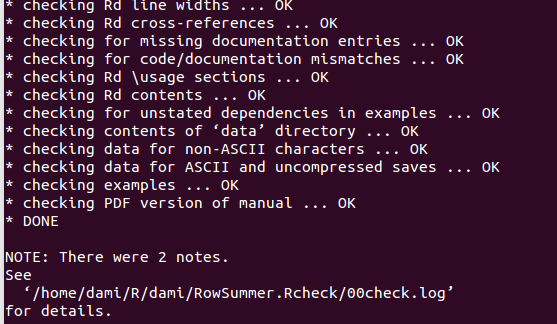
- Now you can check your package by typing in a teminal shell: R CMD check –as-cran PackageName. This may take a while.
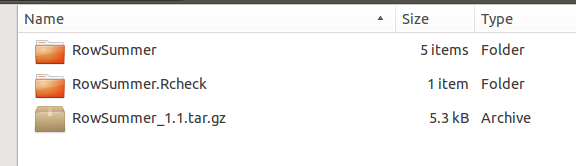
- The CHECK command should return no errors or warnings (check the Notes, if any). CRAN won’t accept packages that return Warnings or Errors during a check. After this operation, the the working directory should contain:
- the folder generated by the package.skeleton() call
- the tarball generated by the R CMD build command (our new package)
- a Rcheck folder containing the files and the logs generated during the check operation
- The package is built and ready to be shared with the R community
Submit an R package to CRAN
The tarball you prepared in the previous step can be directly submitted to CRAN. CRAN will double check the file you are submitting and will contact you with the list of issues that require to be fixed. You can submit your tarball at the following URL: https://cran.r-project.org/submit.html. Before submitting a package, it is important to make sure that the package can be built on Windows R-Release and R-Devel. You can test this by submitting your tarball at the following server: http://win-builder.r-project.org/upload.aspx. You will be informed via email about the results of the test.
Publish an R package on GitHub
I assume that you already have a GitHub account and that you have git installed on your Linux system. If you are new to git, you may start by having a look at this post: https://www.digitalocean.com/community/tutorials/how-to-install-git-on-ubuntu-14-04. I also assume that you just prepared an R package according to the steps described before. I learned the basics of publishing R packages on GitHub from here: http://kbroman.org/pkg_primer/pages/github.html. That’s a very nice post, you should have a look at it.
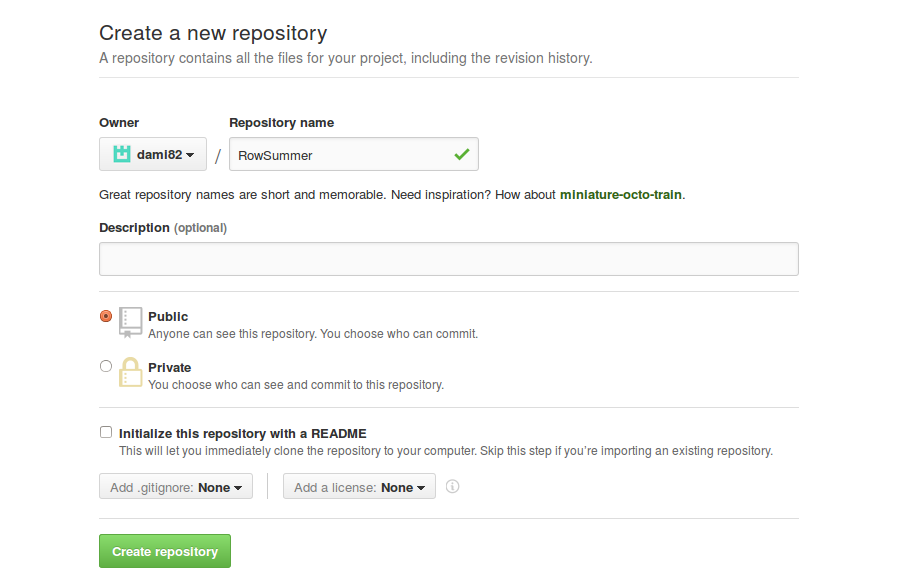
- Log-in to GitHub and create a new repository.
- Once the GitHub repository has been created, you can prepare the local files for the submission
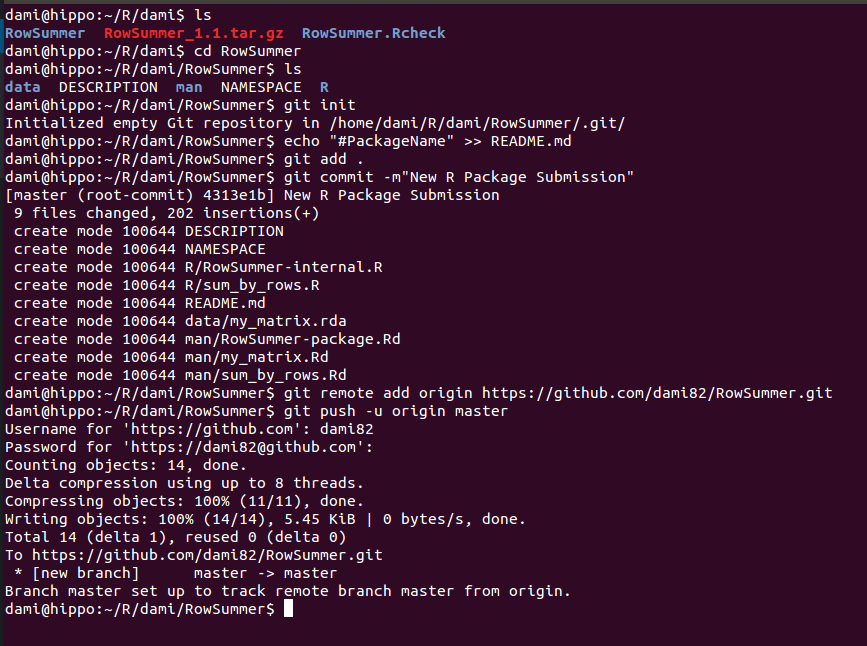
- Open a Terminal shell and change to the directory containing the files generated by package.skeleton().
- Run the following command in the terminal shell: git init. This will initialize an empty repository (/.git) in the local folder
- Generate a README.md file by typing: echo “# PackageName” >> README.md
- Add files to the repository with the command git add . (the point is a regex, it will add all files in the current directory to the repository)
- Write a Commit Message. You can write a one-line message with git commit -m”New R package submission”. It is recommended to write a decent commit message instead of a 1-sentence message. You can do so by typing git commit (this will open Nano in the terminal).
- Invoke the following command: git remote add origin https://github.com/username/RepoName.git (this will connect your local files to the online GitHub repository).
- Finally, push everything to github via git push -u origin master (you can force via git push -uf origin master).
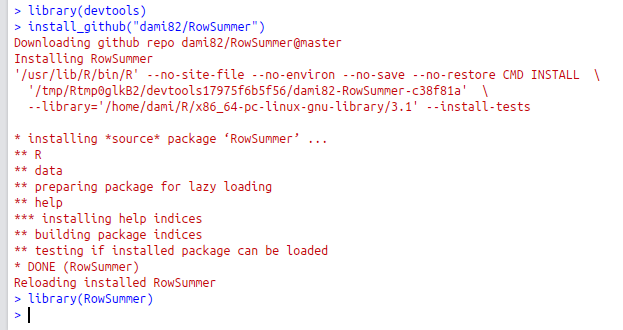
 That’s it! Success! Enjoy! If everything worked as expected, it should be possible to install your package from R using {devtools}.
That’s it! Success! Enjoy! If everything worked as expected, it should be possible to install your package from R using {devtools}.